Note: This story was last updated at 10:55 a.m.
The Summit supercomputer at Oak Ridge National Laboratory has been used to identify drug compounds, including medications and natural compounds, that could help fight coronavirus, although more study is needed.
“The researchers used Summit, the world’s most powerful and smartest supercomputer, to identify 77 small-molecule drug compounds that might warrant further study in the fight against the SARS-CoV-2 coronavirus, which is responsible for the COVID-19 disease outbreak,” ORNL said in a response to questions Wednesday.
The researchers performed simulations on Summit of more than 8,000 compounds to screen for those that are most likely to bind to the main “spike” protein of the coronavirus, rendering it unable to infect host cells. They ranked compounds of interest that could have value in experimental studies of the virus. They published their results on “ChemRxiv.”
Chinese researchers have sequenced the virus, and they discovered that it infects the body by one of the same mechanisms as the Severe Acute Respiratory Syndrome, or SARS, virus that spread to 26 countries during the SARS epidemic in 2003, ORNL said in a press release Thursday. The similarity between the two virus structures helped with the study of the new virus.
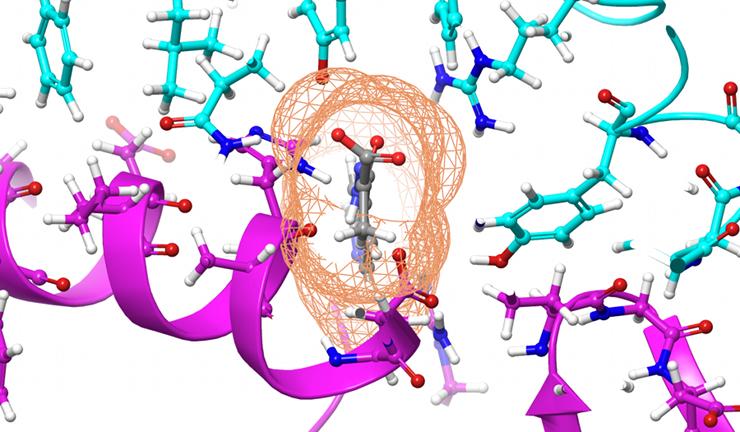
Jeremy C. Smith, Governor’s Chair at the University of Tennessee and director of the UT/ORNL Center for Molecular Biophysics, assumed the two viruses may even “dock” to the cell in the same way, ORNL said. Team member and UT/ORNL CMB postdoctoral researcher Micholas Smith built a model of the coronavirus’ spike protein, also called the S-protein, based on early studies of the structure.
“We were able to design a thorough computational model based on information that has only recently been published in the literature on this virus,” Micholas Smith said, referring to a study published in “Science China Life Sciences.”
After being granted computational time on Summit through a Director’s Discretionary allocation, Micholas Smith used a chemical simulations code to perform molecular dynamics simulations. Those analyze the movements of atoms and particles in the protein. He simulated different compounds docking to the S-protein spike of the coronavirus to determine if any of them might prevent the spike from sticking to human cells, ORNL said.
“Using Summit, we ranked these compounds based on a set of criteria related to how likely they were to bind to the S-protein spike,” Micholas Smith said.
The team found 77 small-molecule compounds, such as medications and natural compounds, that they suspect may be of value for experimental testing. In the simulations, the compounds bind to regions of the spike that are important for entry into the human cell, and therefore might interfere with the infection process, ORNL said.
After a highly accurate S-protein model was released in “Science,” the team plans to rapidly run the computational study again with the new version of the S-protein, the lab said. This may change the ranking of the chemicals likely to be of most use. The researchers emphasized the necessity of testing of the 77 compounds experimentally before any determinations can be made about their usability.
“Summit was needed to rapidly get the simulation results we needed. It took us a day or two whereas it would have taken months on a normal computer,” Jeremy Smith said. “Our results don’t mean that we have found a cure or treatment for the Wuhan coronavirus. We are very hopeful, though, that our computational findings will both inform future studies and provide a framework that experimentalists will use to further investigate these compounds. Only then will we know whether any of them exhibit the characteristics needed to mitigate this virus.”
Computation must be followed by experiment. Computational screening essentially “shines the light” on promising candidates for experimental studies, which are essential for verifying that certain chemicals will combat the virus, according to Jeremy Smith. The use of a supercomputer such as Summit was important to get the results quickly.
Researchers started working on the project during the second week of February, ORNL said. The research is funded by ORNL’s Laboratory Directed Research and Development program and the National Institutes of Health (through the University of Tennessee). The Summit supercomputer is a DOE Office of Science User Facility funded by the DOE Advanced Scientific Computing Research program.
Learn more in this video.

Leave a Reply